proteinogenic 
proteinogenic 
Chemical structure generation for protein sequences as [SMILES] string.
This crate builds on top of purr, a crate providing
primitives for reading and writing [SMILES].
Use the AminoAcid enum to encode the sequence residues, and build a SMILES
string with proteinogenic::smiles:
```rust extern crate proteinogenic;
let sequence = "KGILGKLGVVQAGVDFVSGVWAGIKQSAKDHPNA"; let residues = sequence.chars() .map(|c| proteinogenic::AminoAcid::from_code1(c).unwrap());
let s = proteinogenic::smiles(residues); ```
This SMILES string can then be given in conjunction with cheminformatics toolkits, for instance using OpenBabel to generate a PNG figure:
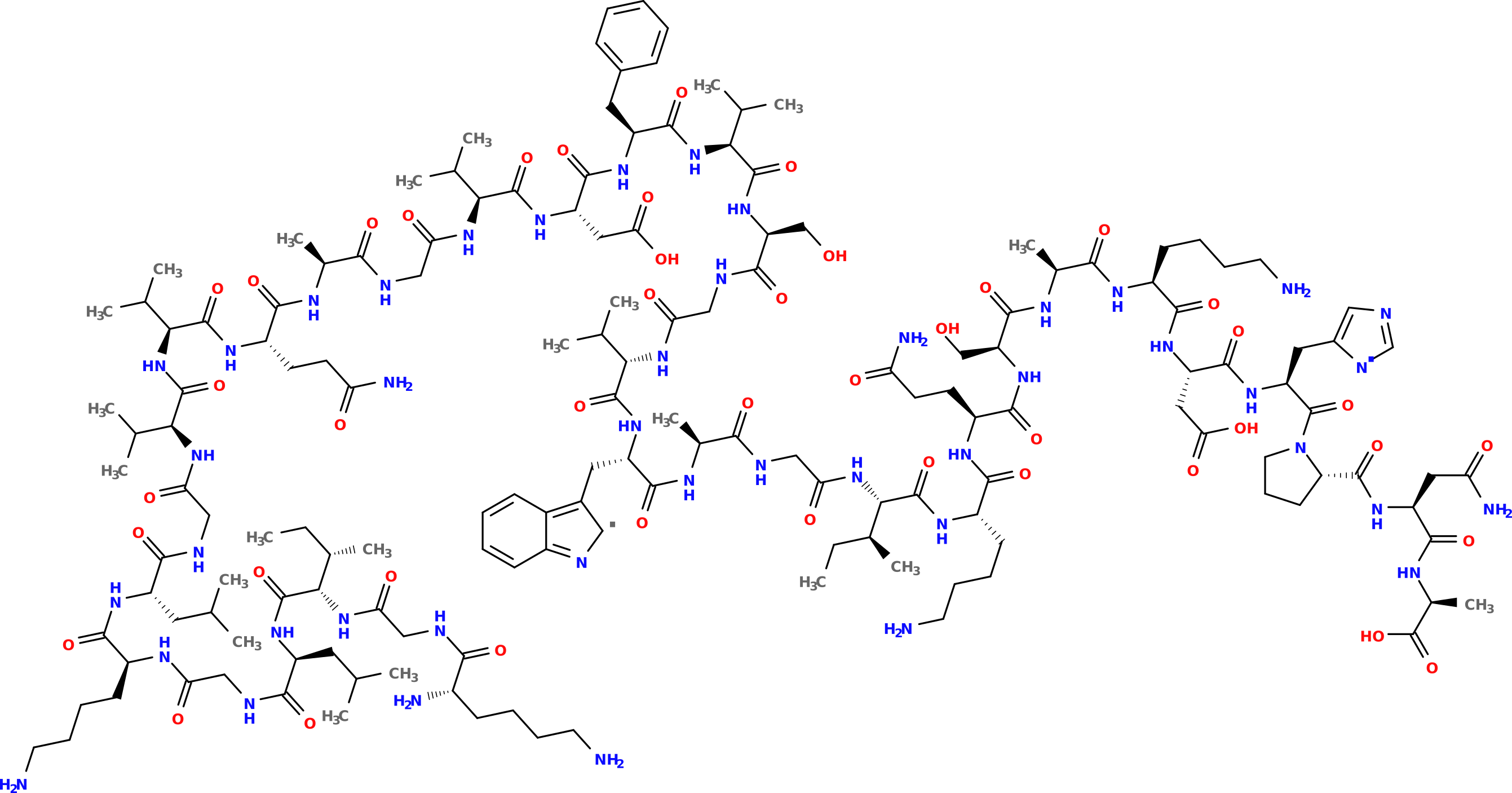
Note that proteinogenic is not limited to building a SMILES string; it can
actually use any purr::walk::Follower
implementor to generate an in-memory representation of a protein formula. If
your code is already compatible with purr, then you'll be able to use
protein sequences quite easily.
```rust extern crate proteinogenic; extern crate purr;
let sequence = "KGILGKLGVVQAGVDFVSGVWAGIKQSAKDHPNA"; let residues = sequence.chars() .map(|c| proteinogenic::AminoAcid::from_code1(c).unwrap());
let mut builder = purr::graph::Builder::new(); proteinogenic::visit(residues, &mut builder);
builder.build() .expect("failed to create a graph representation"); ```
The API is not yet stable, and may change to follow changes introduced by
purr or to improve the interface ergonomics.
Found a bug ? Have an enhancement request ? Head over to the GitHub issue tracker if you need to report or ask something. If you are filing in on a bug, please include as much information as you can about the issue, and try to recreate the same bug in a simple, easily reproducible situation.
This project adheres to Semantic Versioning and provides a changelog in the Keep a Changelog format.
If you're a bioinformatician and a Rustacean, you may be interested in these other libraries:
uniprot.rs: Rust data structures
for the UniProtKB databases.obofoundry.rs: Rust data
structures for the OBO Foundry.fastobo: Rust parser and abstract
syntax tree for Open Biomedical Ontologies.pubchem.rs: Rust data structures
and API client for the PubChem API.This library is provided under the open-source MIT license.
This project was developed by Martin Larralde during his PhD project at the European Molecular Biology Laboratory in the Zeller team.