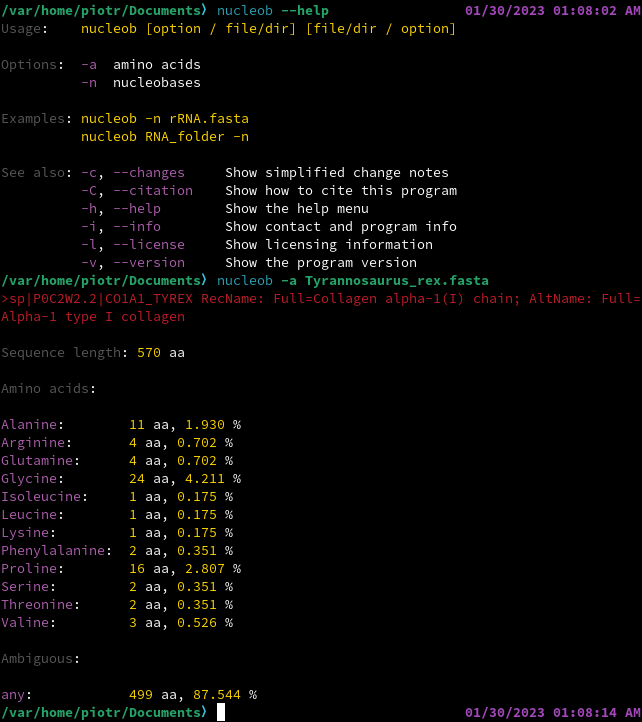
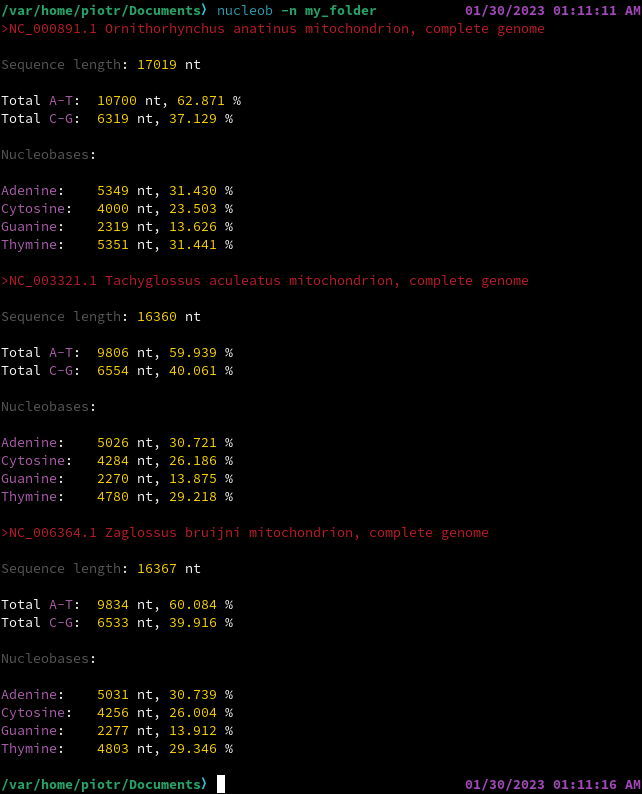
nucleob performs a simple statistical analysis of .fasta files with either nucleotide or amino acid sequences. It is designed to give (1) highly reliable and (2) clearly formatted results, being also (3) supremely minimalist and (4) cross-platform.
[keywords: amino acids, bioinformatics, biology, DNA, fasta, genes, genetics, molecular, nucleic acids, nucleobases, nucleotides, proteins, RNA, scientific computing, statistics, stats]

You can modify and fork nucleob under terms of the MIT License. Citing nucleob in a research article, always refer to a specific program version, e.g.:
Bajdek, P., 2022. nucleob (version 1.1.0). [computer software] https://github.com/piotrbajdek/nucleob
nucleob should run smoothly on Windows and macOS, and can be installed by the use of cargo. Yet, it is being developed and primarily tested on Fedora Linux.
nucleob v1.1.0:
– Was successfully tested on Arch Linux, Fedora Linux 37, openSUSE Tumbleweed, Ubuntu 22.04 and Ubuntu 22.10.
– Failed to run on Mageia 8 due to an old glibc version (required ≥2.34).
[recommended for programmers]
1. Install from crates.io by the use of cargo:
cargo install nucleob
By default, the file will be downloaded to .cargo/bin/, a hidden folder in your home directory.
2a. For convenience, you will probably want to copy nucleob to /usr/bin/ as in Method 2 (3a, 3b).
2b. Alternatively, add ~/.cargo/bin directory to your PATH variable (can be set up by rustup).
1. Download the distro-independent binary of nucleob from GitHub.
2. Make the file executable:
sudo chmod +x ./nucleob
3a. On most Linux distros, install nucleob via copying the binary to /usr/bin/:
sudo cp nucleob /usr/bin/
3b. On Fedora Silverblue / Kinoite:
sudo cp nucleob /var/usrlocal/bin/
[recommended for most users]
Distro-specific packages are also available for download for .rpm- and .deb-based Linux distros. Installation instructions:
Fedora Linux / RHEL / openSUSE:
sudo rpm -i nucleob-1.1.0-1.x8664.rpm_
Fedora Silverblue / Kinoite:
rpm-ostree install nucleob-1.1.0-1.x8664.rpm_
Ubuntu:
sudo dpkg -i nucleob1.1.0amd64.deb
Download and unpack the nucleob source from GitHub. Then, build and install the program:
cargo build --release && sudo cp target/release/nucleob /usr/bin/